Biomass Report 8/1/2025
Report Overview
Recent field surveys and drone flights at the US-SRG flux site enabled an exploration of woody biomass estimation methods at fine and coarse spatial scales. In this report, we estimated woody biomass using three approaches:
- Allometric equations based on field-measured basal diameter
- Allometric equations applied to canopy metrics
- Voxel-based calculations using high-density LiDAR
In addition to estimating total biomass at the site, we apply these methods to estimate biomass along the biometric mesquite gradient at US-SRG. Finally, we examined trends in biomass products available at the site.
Allometry with Basal Diameter
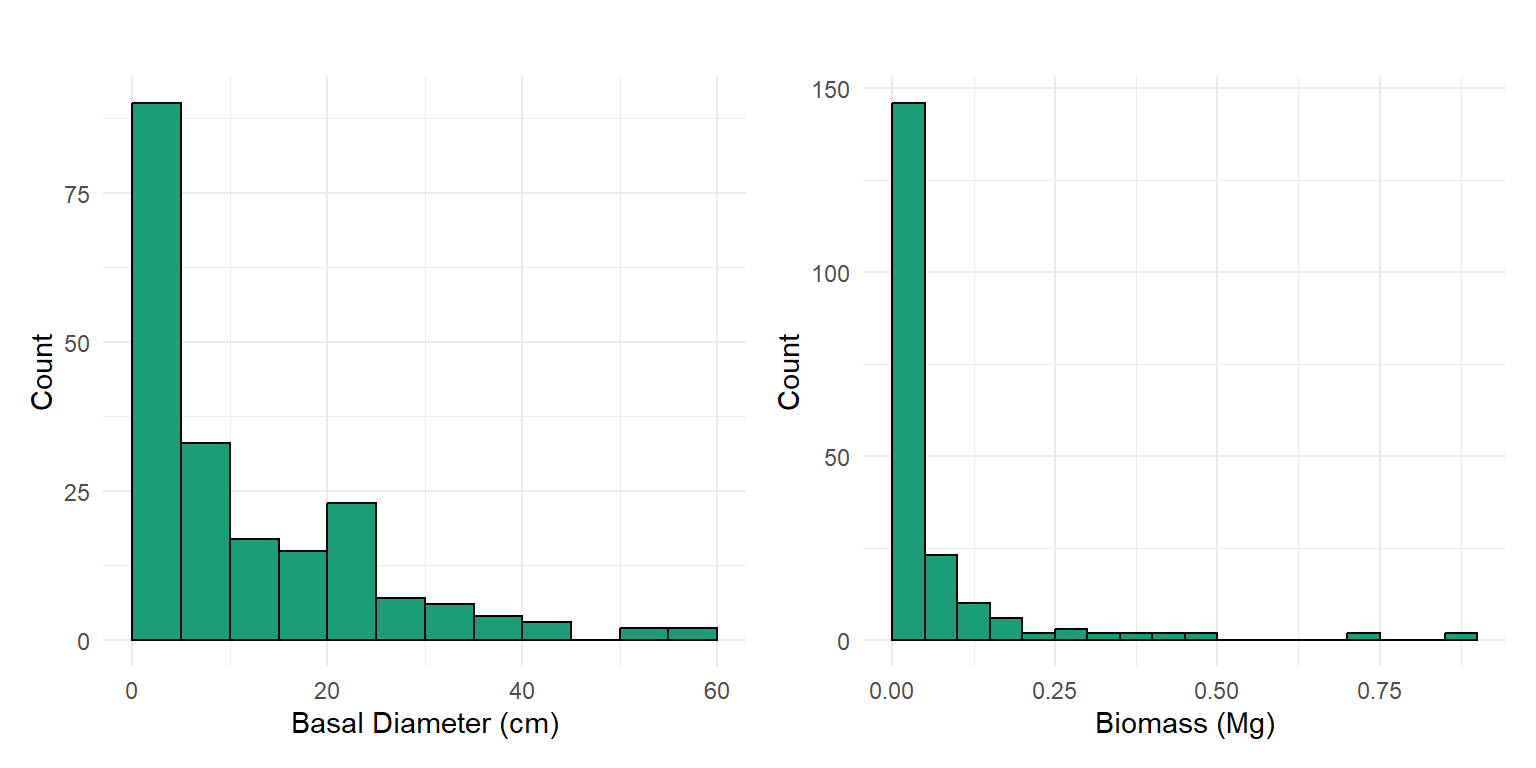
At the end of May 2025, we conducted a woody plant census within a 100-meter radius of the US-SRG flux tower. During this survey, we recorded basal and canopy diameter for all individual shrubs and trees. To estimate aboveground biomass, we applied an allometric equation originally described by Jenkins et al. (2003) and later revised by Chojnacky et al. (2013). Given that the vegetation at US-SRG is dominated by mesquite (Prosopis spp.), we used the equation specific to Woodland Fabaceae/Rosaceae:
Biomass = exp(−2.9255 + 2.4109 × log(x))
where x is diameter at root collar (DRC) in centimeters.
We made two key assumptions in applying this relationship. First, because DRC was not measured directly, we approximated it using basal diameter measurements. Second, although the equation is primarily based on mesquite data, we include additional woody desert shrubs: hackberry (Celtis pallida), greythorn (Ziziphus obtusifolia), whitethorn acacia (Vachellia constricta), and catclaw acacia (Senegalia greggii). For multi-stemmed individuals, we used the average basal diameter across stems.
For desert willow (Chilopsis linearis), we applied a separate equation using diameter at breast height (DBH), with parameters as follows:
Biomass = exp(−2.4441 + 2.4561 × log(x))
where x is DBH in centimeters.
The total woody biomass from the census basal diameter observations is 12.66 Mg.
Allometry with Canopy Metrics
In order to estimate biomass from canopy metrics, we modeled an empirical relationship between canopy area and woody biomass using observations from the biometric gradient:
Biomass = 0.1136 * (Canopy Area)^1.8890
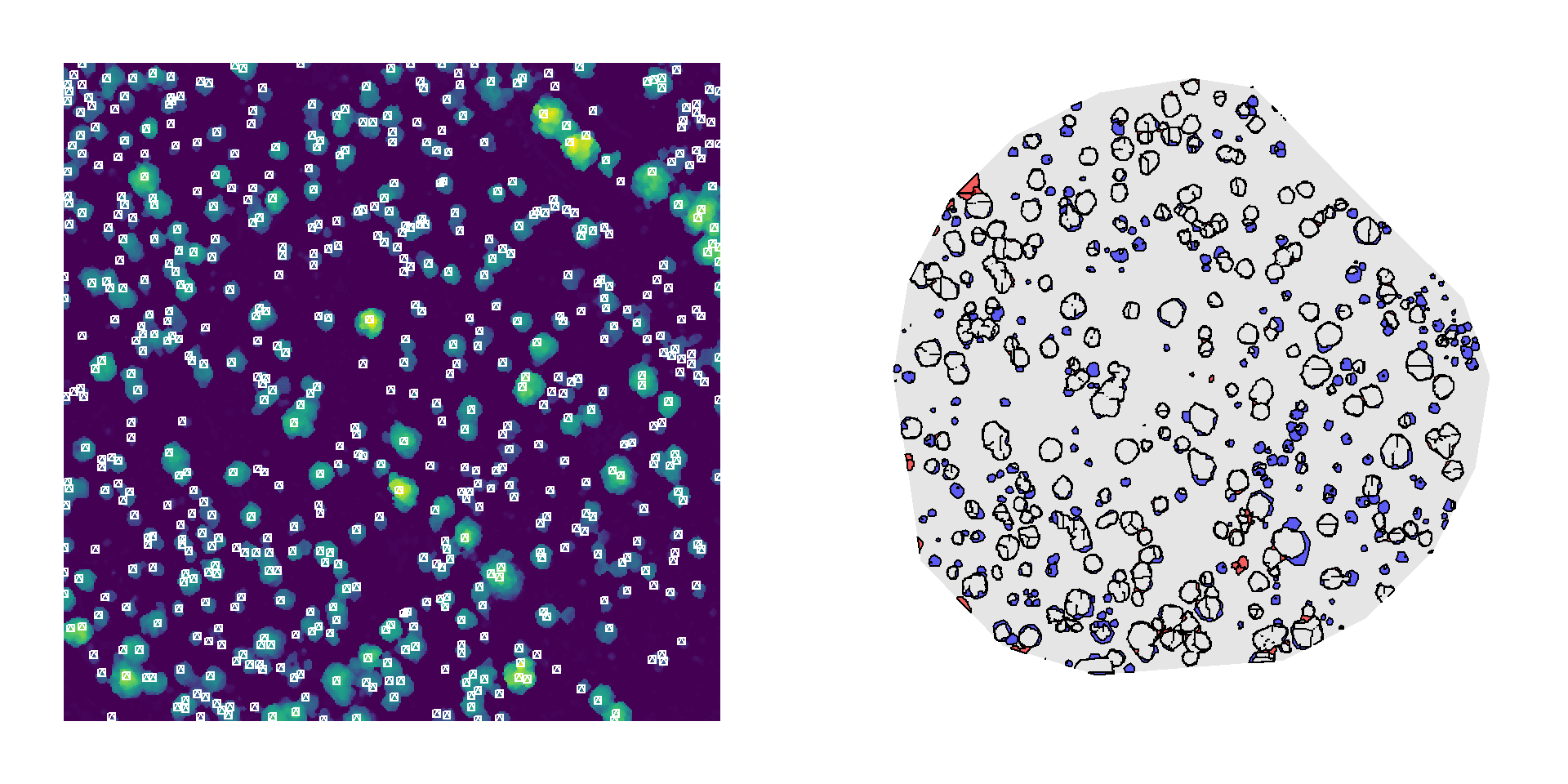
Canopy area was derived automatically from a LiDAR-based canopy height model (CHM) using treetop detection and watershed segmentation to delineate individual tree crowns. We then applied the canopy area–biomass relationship to estimate total woody biomass within a 100-meter radius of the US-SRG flux tower for the automatically detected crowns, from manually detected crowns (see DronetoCensusComparison report in FluxandtheField repository), and crown metrics from field observations.
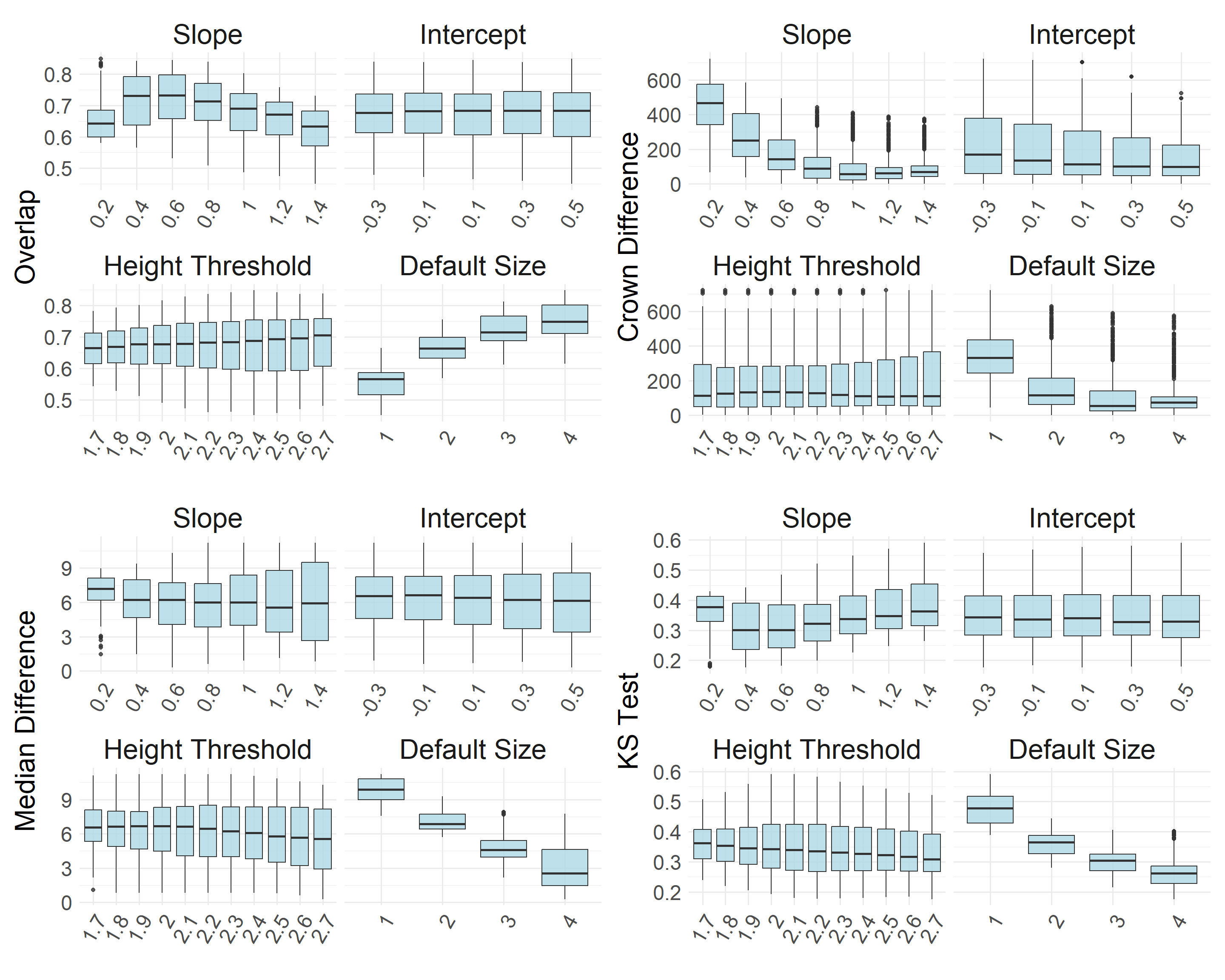
To detect treetops on the CHM, we used a variable window filter, where the window size is a linear function of canopy height (y = ax + b). In the function, we also include a height threshold below which a default window size is applied. To identify optimal parameters for this function, we evaluated over 1,500 combinations of values based on their ability to identify treetops and create a reasonable distribution of canopy areas relative to the manually delineated canopy crowns. More specifically, we evaluated the parameters based on the outputs’:
Deviation in the number of detected crowns compared to manual delineation
Deviation in the median canopy area
Deviation in the distribution of canopy areas
Statistical difference in distribution shape (Kolmogorov–Smirnov test)
The optimal window function based on the parameters tested was:
Window size = 0.6x + 0.3 Window height threshold: < 2.6 m, window size = 4 m
This combination of parameters agreed strongly with the manually delineated canopy area metrics, resulting in a crown count difference of 1, median canopy area difference of 1.45 m2, an overlap score of 0.82, and a Kolmogorov–Smirnov D value of 0.22.
Although this method occasionally overdetects crowns in taller vegetation (left), and the segmented canopy polygons often underestimate crown area even when treetops are correctly located (right), the overall performance is comparable to the manually delineated reference.
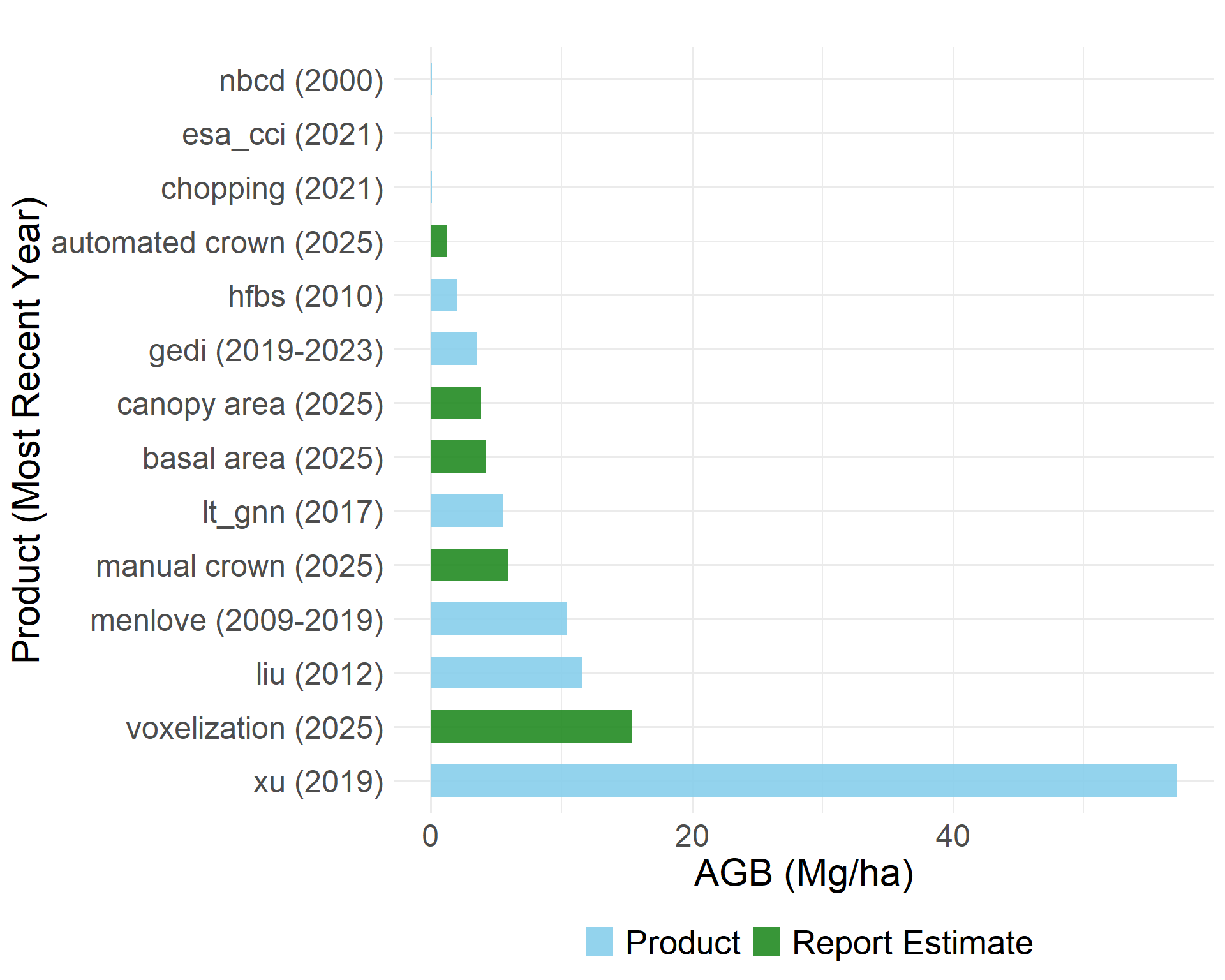
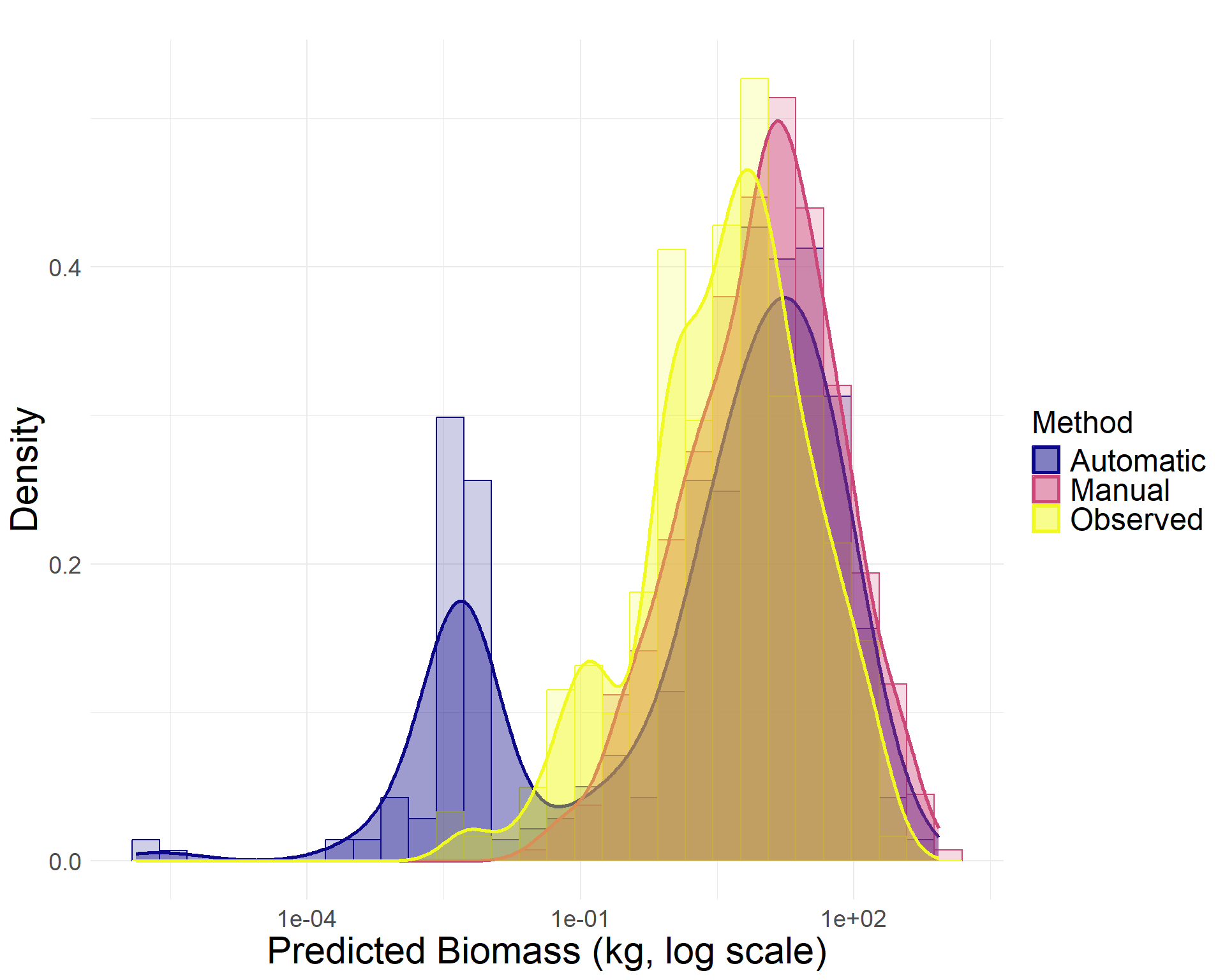
To further assess precision, we compared total woody biomass estimates derived from three sources of canopy area measurements:
Automated LiDAR-based segmentation: 11.7 Mg
Manual canopy delineation: 18.0 Mg
Field observations: 3.8 Mg
Voxelization
For the final biomass estimation method, we used voxelized a LiDAR point cloud data a cube size of 2.5 cm. Points were filtered to exclude noise and low-returns associated with grasses. Unlike the segmentation-based method, voxelization allowed us to estimate biomass across the entire area without requiring delineation of individual tree crowns.
We applied a specific wood density of 0.78 g cm-3 (Chojnacky et al., 2013), to convert voxelized volume into woody biomass. Using this method, the total estimated biomass within the survey boundary was 46.9 Mg.
It is important to note, however, that this method is highly sensitive to voxel resolution. For example, increasing the voxel side length to 5 cm resulted in a biomass estimate of 325.6 Mg. Further work is needed to understand the influence of voxel resolution on estimating biomass and what criteria is needed to select a voxel size.
Biometric Gradient Comparison
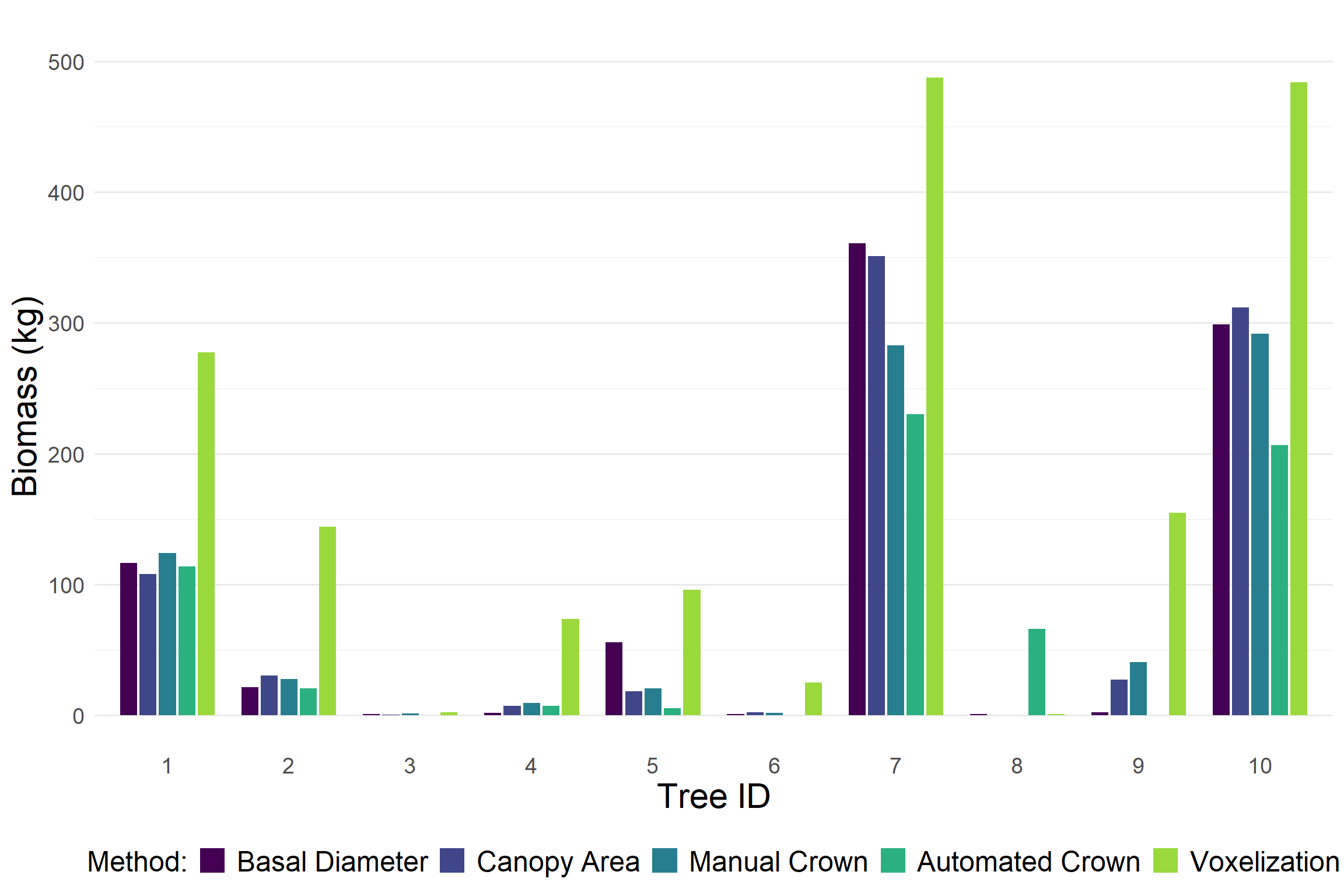
After evaluating biomass estimation methods at the ecosystem scale, we narrowed our focus to a set of 10 individual trees spanning a wide range of heights, basal diameters, canopy diameters, and growth forms. We refer to these tree as the biometric gradient. For each tree, we estimated aboveground biomass using all available methods described above.
Voxel-based estimates consistently produced higher biomass values compared to other approaches. In contrast, the automated crown detection method tended to output lower biomass estimates. The estimates from basal diameter and canopy diameter were generally similar. This was expected since both rely on the same allometric relationship.
Biomass Products
Direct comparison between the biomass estimates from the methods explored above and existing biomass products is challenging due to the differences in temporal coverage and original spatial resolution. We compare them regardless and observe substantial variability across both fine and large scale approaches.